Abstract
Introduction The application of circulating cell-free DNA (cfDNA) has attracted recent interest for cancer detection and monitoring. The presence of somatic copy number alterations (SCNAs) can be used to differentiate between cancer cases and healthy controls (PMID: 29109393). This information can be retrieved from low-coverage whole genome sequencing (lcWGS) and used to determine the proportion of tumor derived cfDNA (ctDNA) in a tube of blood at a reasonable cost.
Previous studies observed that cancer patients tend to have a higher fraction of shorter cfDNA fragments compared to healthy controls, irrespective of the presence of genomic events (PMID: 30404863).
Here, we evaluate if a combination of cfDNA genomic and epigenomic features that can be retrieved from a single run of lcWGS, has potential for the early prediction of end of induction (EoI) response, in patients with high-grade lymphoma with MYC and BCL2 and/or BLC6 rearrangements (HGBL-DH/TH).
Methods In the HOVON-152 phase II trial (NCT03620578) HGBL-DH/TH patients were treated with induction immunochemotherapy with one cycle of R-CHOP followed by five cycles of DA-EPOCH-R. Patients in complete metabolic response (CMR, defined as Deauville score 1-3) at the end of induction receive 1-year nivolumab consolidation. Blood was collected from 40 patients using PAXgene ccfDNA tubes after one cycle (timepoint 1, T1), three (T2) and six (T3) cycles of treatment. At T3, response to induction treatment was determined using 18F-FDG-PET scan. Patients with CMR at EoI were considered responders. In this study, we included 40 out of 97 patients from the HOVON-152 phase II trial, and enriched for patients with no CMR. Plasma was separated using dual centrifugation protocol, and cfDNA was isolated using the QIASymphony kit (QIAGEN). Sequencing libraries were prepared using the ThruPLEX Plasma-seq kit (Takara). lcWGS was performed on a Novaseq 6000 (Illumina). Somatic copy number aberrations (SCNAs) were retrieved with ichorCNA, insert size profiles were generated using Picard tools. SCNAs were classified as detected when the tumor fraction quantified with ichorCNA exceeded 3%.
Results We assessed the detection of SCNAs and the SCNA-derived tumor fraction using the ichorCNA software. The ctDNA tumor fraction was found to be significantly increased in non-responders (NR, n=24) compared to responders (R, n=16), irrespective of sampling timepoint (Wilcoxon, p<0.001). At T1, the tumor fraction was significantly elevated in non-responders (Wilcoxon, p=0.014), and SCNAs were detected in 6/16 (38%) of the responders versus 19/24 (79%) of the non-responders, with negative predictive value (NPV): 71%, positive predictive value (PPV): 74%. Therefore, non-responders tend to have a higher fraction of tumor-derived signal which is already apparent at earlier timepoints.
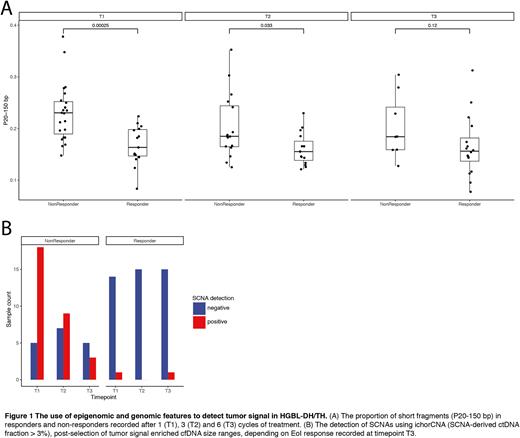
To evaluate the potential of using cfDNA fragmentation for the early detection of response, we retrieved the fragment size profiles. The proportion of short fragments (P20-150 bp), was found to be increased in non-responders compared to responders at T1 (Wilcoxon, p<0.001) and T2 (Wilcoxon, p=0.033), which suggests that the plasma of non-responders contains a higher fraction of tumor-signal (Figure 1A).
Previous reports leveraged the size properties of ctDNA in order to increase the sensitivity of SCNA based methods (PMID: 30404863). In order to utilize both epigenomic and genomic properties to increase the detection rate of tumor-signal, we performed in silico size selection to computationally filter out fragments in the 20-150bp size range. Upon repeating the SCNA analysis, we observed that 20/24 (83%) non-responders and 1/15 (19%) responders had detectable ctDNA at T1 (Figure 1B). This yields a NPV and PPV of 80% and 86% respectively for EoI response detection at T1. By leveraging the fragmentomic properties of cfDNA, the sensitivity of SCNA-based methods can be improved, which leads to an increased detection of response at earlier timepoints.
Discussion In patients with HGBL-DH/TH, after one cycle of R-CHOP, the presence of SCNAs enhanced by fragment size analysis in plasma, is significantly associated with an unfavourable EoI response, with a NPV and PPV of 80% and 86% respectively. This novel approach can be performed without prior knowledge of genetic alterations, and may have high potential to guide early risk-adapted treatment strategies in HGBL-DH/TH.
Disclosures
Nijland:Roche: Research Funding; Takeda: Research Funding; Genmab: Consultancy. Klerk:Roche: Current Employment, Honoraria. Vergote:Beigene: Consultancy; Celgene/BMS: Consultancy; Gilead/Kite: Consultancy; Janssen: Honoraria. Pegtel:ExBiome: Current equity holder in private company; Amgen: Research Funding; Takeda: Consultancy, Research Funding; Gilead: Research Funding. Chamuleau:Gilead: Research Funding; Roche: Honoraria; Genmab: Research Funding; Abbvie: Honoraria; Novartis: Honoraria; BMS/Celgene: Honoraria, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal